Bridging Genome to Function: Post-Genome Science
Learning Objectives
- Identify potential ontology databases
- Analyze functional pathways of:
- ChIP-Seq
- DEG
- Describe basic protein-protein interaction networks
Functional View in Genomics
Connecting Differences to Pathways
Motivation
- If 300 genes are differentially expressed, what kind of biological pathways are affected?
- What are the implications of DNA-microarray/ChIP-seq results in the clinical perspective?

- I cannot lose weight because my genes involved in fat synthesis are upregulated compared to others.
Need for an Encyclopedia
- We have databases to annotate each gene with its:
- Location in the genome
- Molecular function
- Biological pathways it involves
- Co-regulated/expressed genes
- Properties
Databases Recording All Information
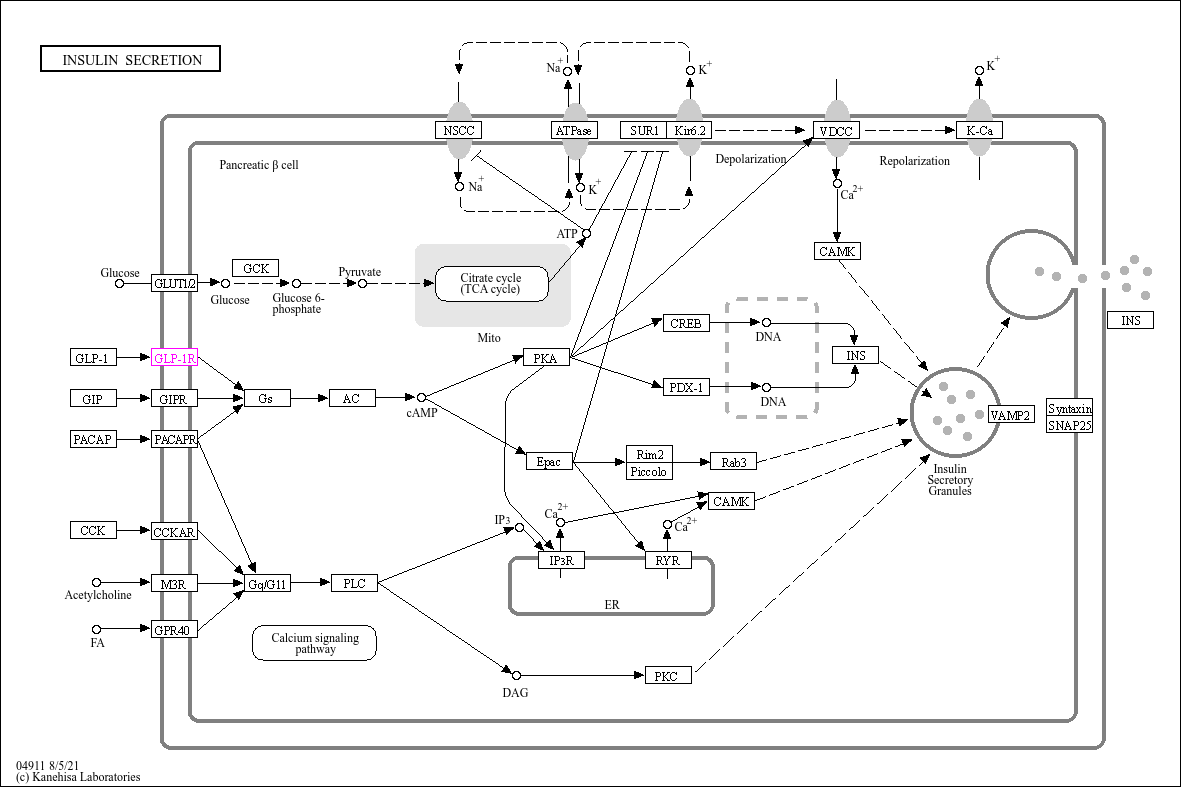
Kyoto Encyclopedia of Genes and Genomes (KEGG)
- Integrates genomic, biological pathway, disease, and drug information
- Pathway maps depict molecular interaction networks including metabolic, signaling, and genetic information pathways.
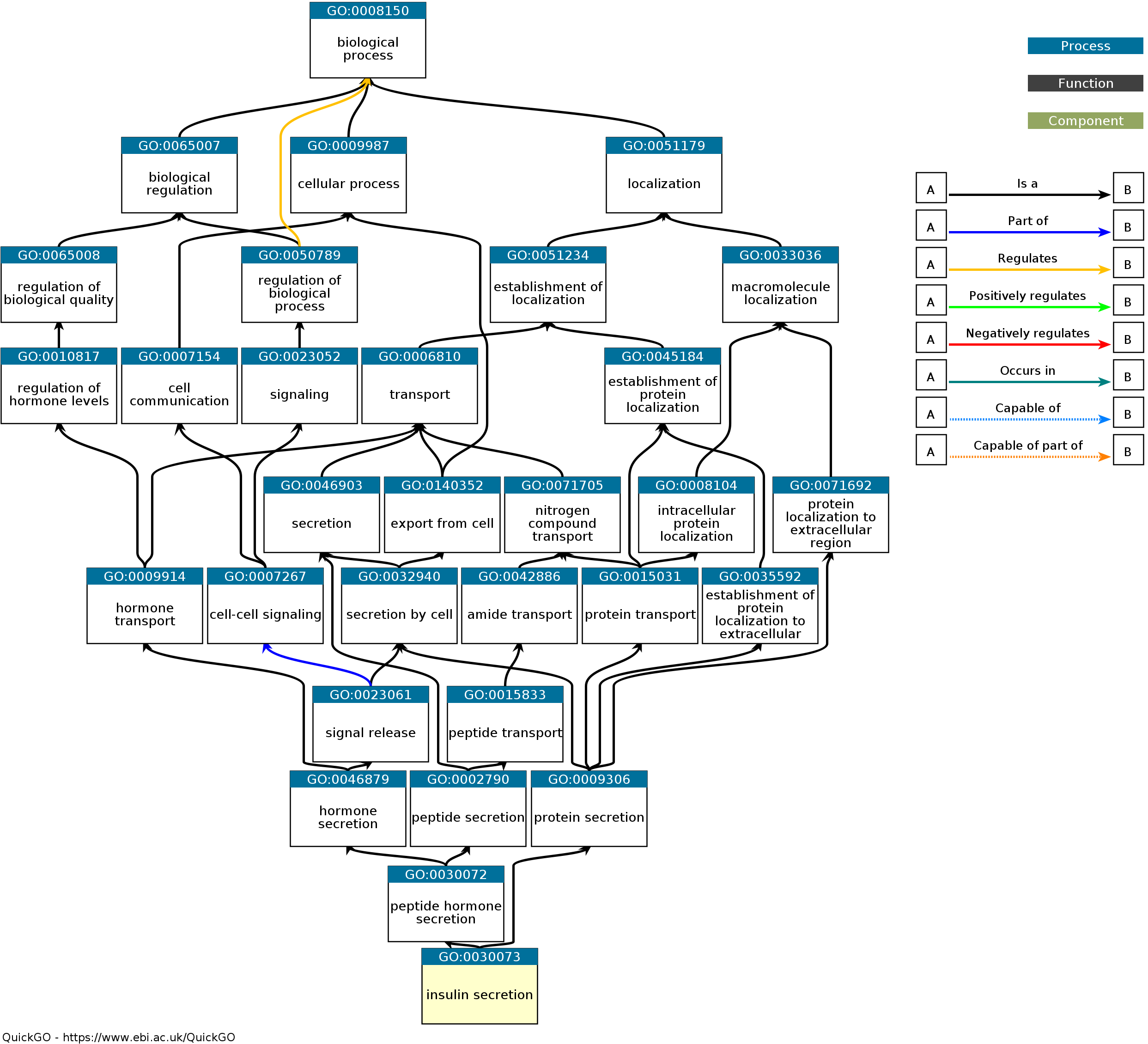
Gene Ontology (GO)
- GO is a standardized, hierarchical ontology for annotating gene products across species
- Annotates from three perspectives:
- Biological process
- Cellular component
- Molecular functions
- Biological process
Example of KEGG Pathways
Example of GO Pathways
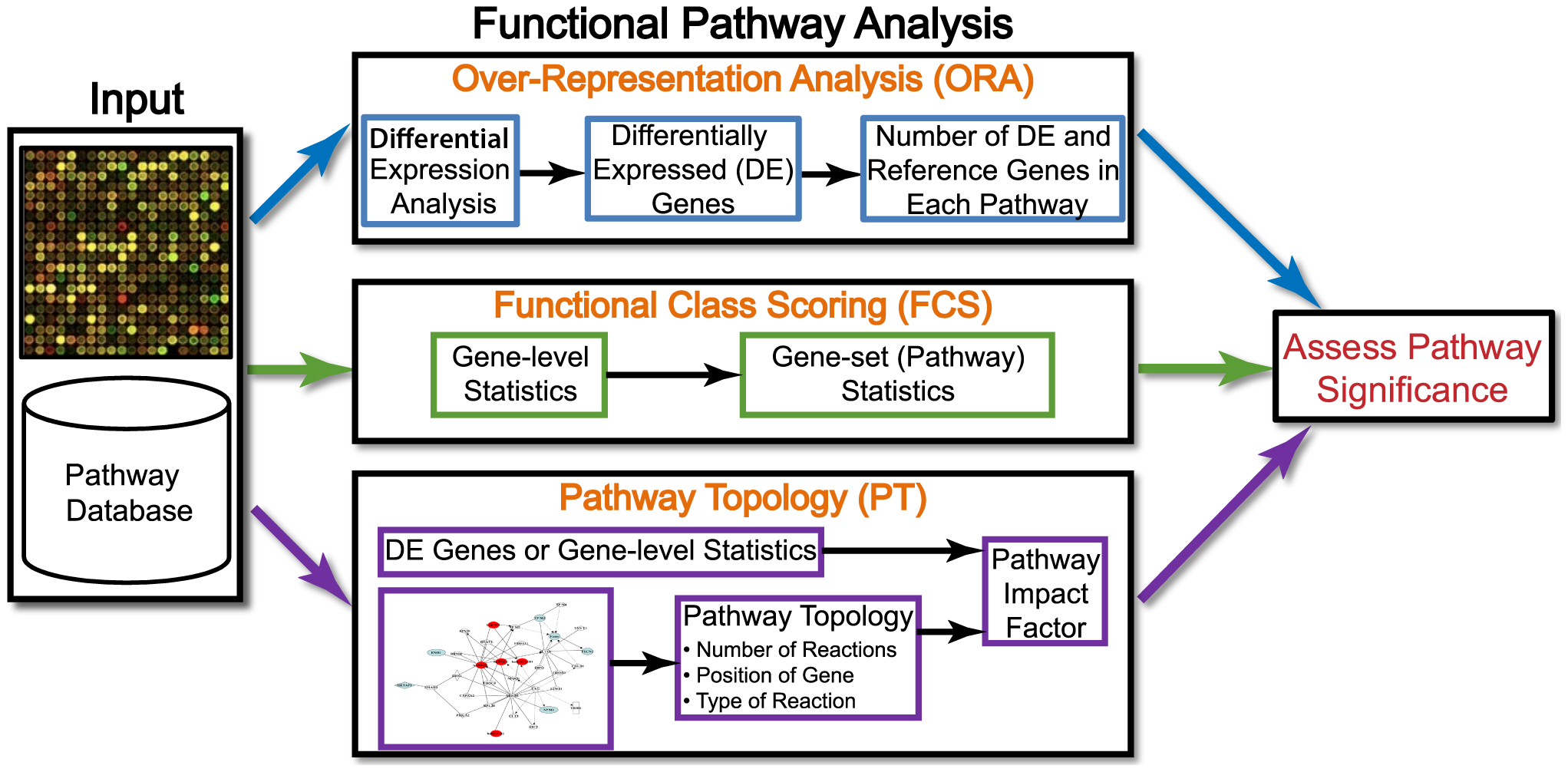
Enrichment Analysis
Connecting Genes to Biological Functions
Enrichment Analysis Summarizes identified differences in genes associated with common functions.
Enrichment in the Sequence
Genomic Regions Enrichment of Annotations Tool
- Motivation: Do cis-regulatory elements associate with specific pathways?
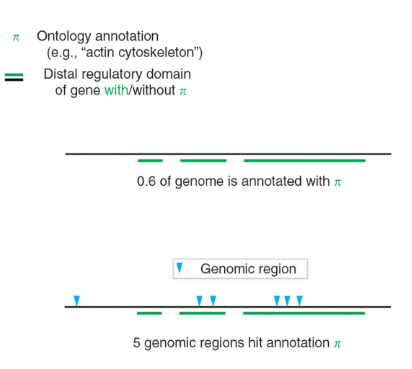
Problem Formulation
- Given a 1000 bp sequence
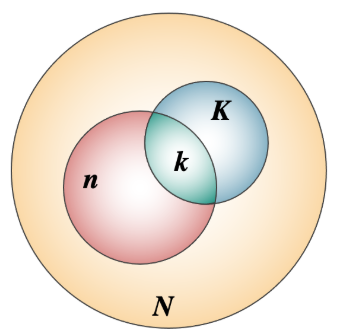
- From the existing database, 600 bp (located in the figure) is annotated with the functional path \(\pi\)
- ChIP-seq identified 6 enriched regions bound by protein, among them, 5 are in the annotated region \(\pi\)
- Question: If the ChIP-Seq identified regions occur randomly, what is the probability that ChIP-Seq identified regions fall into the regulatory domain \(\pi\)?
Binomial Test
Analogy: You are flipping an unfair coin
- Chance of having a head: 0.6
- Times you flip: 6
- Heads you get: 5
ChIP-seq: Identified Region
- Chance to be in the regulatory domain: \(p = \frac{\text{Annotated length}}{\text{Total ChIP-seq length}} = \frac{600}{1000}\)
- Number of genomic regions identified: \(n = 6\)
- Number of genomic regions falling into the annotated area: \(k = 5\)
The probability of getting exactly k heads out of n with the probability \(p\):
\[ P(X = k) = \binom{n}{k}(p)^k(1-p)^{n-k} \]
Back to Our Example
- We can derive the p-value (probability) to test: How likely is it to get at least \(k\) annotations in the regulatory region out of \(n\) attempts, if the ChIP-Seq peaks occur randomly?
\[ \text{p-value} = P(X \geq k) = \sum_{i=k}^{n}\binom{n}{i}(p)^i(1-p)^{n-i} \]
In our example, it will be
\[ P(X\geq 5) = \binom{6}{5}(0.6)^5(1-0.6)^{1} +\binom{6}{6}(0.6)^6(1-0.6)^{0}=0.23 \]
For a given false discovery rate (FDR, typically 0.05), if \(\text{p-value} < FDR\), we will say that the regulatory element is enriched in the pathway \(\pi\).
Wrap-up: Enrichment Analysis of a Set of Cis-Regulatory Regions
For a sequence with length \(N\) with \(K_\pi\) of them annotated with pathway \(\pi\)
ChIP-seq selected \(n\) genomic regions and \(k_\pi\) of them fall into the annotation of pathway \(\pi\)
\(p_\pi\) is the probability of selecting a base pair annotated with \(\pi\) when selecting a single base pair uniformly from all base pairs in the genome \[ \text{p-value} = P(X \geq k_\pi) = \sum_{i=k_\pi}^{n}\binom{n}{i}(p_\pi)^i(1-p_\pi)^{n-i} \]
Each pathway needs to be tested once, and when testing on more than one pathway, P-value correction (e.g., Bonferroni or other methods) should be applied.
Exercise Question
- On a sequence with length \(100\), with \(14\) of them annotated with the pathway multicellular organismal development
- ChIP-seq on this sequence showed that 10 enriched regions are bound by protein in older adults compared to young adults; among them, 6 fall into the annotated regions
Q1 What is the p-value of the binomial test that at least 6 out of 10 fall into the annotated regions?
\[ P(X\geq 6) = \sum_{i=6}^{10}\binom{10}{i}(0.14)^i(1-0.14)^{10-i} = 0.00095 \]
Q2 Given the p-value, is the protein enriched in the pathway multicellular organismal development at the 0.05 false discovery rate?
Enrichment in the Protein
Over-Representation Analysis
- Motivation: Are biological pathways enriched (over-represented) in an experimentally-derived gene list, e.g., a list of differentially expressed genes (DEGs)?
Problem Formulation
Problem Statement
- 10,000 (\(N\)) genes detected in a microarray study
- 100 (\(M\), G1-G4,G10001~G10096) of them belong to pathway \(\pi\)
- 50 (\(n\)) genes were differentially expressed
| DE | NOT DE | Row sum | |
|---|---|---|---|
| In \(\pi\) | 4(a) | 96(b) | 100 (\(M\)) |
| NOT in \(\pi\) | 46(c) | 9854(d) | 9900 (\(N-M\)) |
| Column sum | 50 (\(n\)) | 9950 (\(N-n\)) | 10000 (\(N\)) |
| Gene | Log2FC | Adjusted p-value |
|---|---|---|
| G1 | -2.8 | 0.010 |
| G2 | 2.6 | 0.028 |
| G3 | 2.5 | 0.031 |
| G4 | 2.4 | 0.040 |
| \(\vdots\) | \(\vdots\) | \(\vdots\) |
| G9999 | 0.1 | 0.849 |
| G10000 | 0.4 | 0.733 |
- Missing pathways we can assume they are not DE
Fisher’s Exact Test
- If \(a:c \geq b:d\), then the gene set is enriched with pathway \(\pi\)
- The observation of cells follows a hypergeometric distribution
- Fisher’s exact test (as one of the cells is less than 5)
- The p-value is the probability of the table being more uneven than the observation
\[ P = \sum_{i=a}^M \frac{\binom{M}{i}\binom{N-M}{n-i}}{\binom{N}{n}} = 1- \sum_{i=0}^{a-1} \frac{\binom{M}{i}\binom{N-M}{n-i}}{\binom{N}{n}} \]
Back to Our Example
\[ P = \sum_{i=4}^{100}\frac{\binom{100}{i}\binom{9900}{50-i}}{\binom{10000}{50}} = 1- \sum_{i=0}^{3} \frac{\binom{100}{i}\binom{9900}{50-i}}{\binom{10000}{50}} = 0.0015 \]
- At 5% FDR, the differentially expressed gene is enriched in the pathway \(\pi\)
| Differentially Expressed | NOT Differentially Expressed | Row Sum | |
|---|---|---|---|
| In the pathway \(\pi\) | 4 | 96 | 100 |
| NOT in the pathway \(\pi\) | 46 | 9854 | 9900 |
| Column Sum | 50 | 9950 | 10000 |
Wrap-up: ORA
- Background gene list with size \(N\),
- Pathway gene list \(M\)
- Within the background gene list, \(n\) are differentially expressed and \(a\) of them are in the pathway
\[ P = \sum_{i=a}^M \frac{\binom{M}{i}\binom{N-M}{n-i}}{\binom{N}{n}} = 1- \sum_{i=0}^{a-1} \frac{\binom{M}{i}\binom{N-M}{n-i}}{\binom{N}{n}} \]
- Each pathway needs to be tested once, and when testing on more than one pathway, P-value correction (e.g., Bonferroni or other methods) should be applied.
Pitfalls of ORA
- Arbitrary cutoff of the interested gene list
- Cannot determine the regulation direction:
- Downregulation
- Upregulation
- Even deeper consideration:
- Gene expressions are not independent events
Exercise Question
Consider a genome with 30 genes, where the functional pathway “PATH_X” contains 5 genes {G1, G2, G3, G4, G5}. 10 are differentially expressed genes in males, with Log2 fold change (Log2FC) and adjusted p-values as follows
Q1 Derive the contingency table for the study above to conduct over-representation analysis. use \(p<0.05\) as selection criteria
Q2 What is the p-value for the over-representation analysis
| Gene | Log2FC | Adjusted p-value |
|---|---|---|
| G1 | -2.2 | 0.049 |
| G2 | -2.3 | 0.049 |
| G3 | -2.8 | 0.010 |
| G4 | 2.3 | 0.049 |
| \(\vdots\) | \(\vdots\) | \(\vdots\) |
| G30 | 2.4 | 0.033 |
Solution
Q1
| DE | NOT DE | |
|---|---|---|
| In the pathway | 4 | 1 |
| NOT in the pathway | 6 | 19 |
Q2
\[\begin{aligned} P & = \sum_{i=4}^5 \frac{\binom{5}{i}\binom{25}{10-i}}{\binom{30}{10}} \\ & = \frac{\binom{5}{4}\binom{25}{10-4}}{\binom{30}{10}}+ \frac{\binom{5}{5}\binom{25}{10-5}}{\binom{30}{10}} =0.03124 \\ & = 1-\sum_{i=0}^3 \frac{\binom{5}{i}\binom{25}{10-i}}{\binom{30}{10}} \\ & = 1 - \biggl( \frac{\binom{5}{0}\binom{25}{10-0}}{\binom{30}{10}}+ \frac{\binom{5}{1}\binom{25}{10-1}}{\binom{30}{10}} \\ &+ \frac{\binom{5}{2}\binom{25}{10-2}}{\binom{30}{10}}+ \frac{\binom{5}{3}\binom{25}{10-3}}{\binom{30}{10}}\biggr) =0.03124 \end{aligned} \]
Functional Class Scoring
- Motivation: Genes in the same pathway are usually co-expressed together, but they do not necessarily pass the DEGs threshold selection.
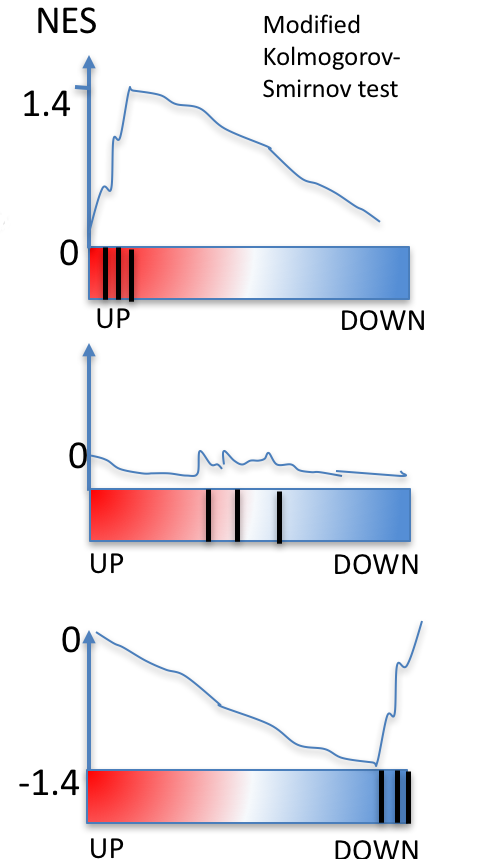
Gene Set Enrichment Analysis (GSEA)
- Sort genes (by log fold change)
- Calculate running sum from the sorted gene list:
- Increase while in the pathway
- Decrease while not in the pathway
- Find the maximum running sum
- Permute the label to calculate p-value
Calculate Running Sum
Consider our gene set with size \(N\), and the pathway \(\pi\) has \(M\) genes in it.
Initial running sum of enrichment score is \(ES_0 = 0\), the denominator \(N_R=\sum |r_j|\), where \(r_j\) is the log fold change of genes that are in the pathway \(\pi\)
Going through the ordered gene \(n_i\) list, do:
- If gene \(n_i\) is in the list, \(ES_i = ES_{i-1} + \frac{|r_j|}{N_R}\)
- If gene \(n_i\) is NOT in the list, \(ES_i = ES_{i-1} - \frac{1}{N - M}\)
The final enrichment score is \(ES = \max(|ES_i|)\)
If DEG missed some genes in the Path !? :
- As long as \(N>M\) you can still use the same formula.
Calculating P-values
Permutation Test: Randomly shuffle the gene label for each expression level for \(k\) times and plot the \(ES\) distribution for these \(k\) permutations, and the p-value will be
\[ \small \text{P-value} = \frac{\text{# of times} ES \geq ES_o}{k} \]
| Gene | Log2FC |
|---|---|
| G1 | 2.3 |
| G2 | 0.7 |
| G3 | 2.6 |
| G4 | 1.4 |
| G5 | 1.2 |
| G6 | 0.1 |
| Gene | Log2FC |
|---|---|
| G4 | 2.3 |
| G2 | 0.7 |
| G6 | 2.6 |
| G1 | 1.4 |
| G5 | 1.2 |
| G3 | 0.1 |
| Gene | Log2FC |
|---|---|
| G4 | 2.3 |
| G1 | 0.7 |
| G6 | 2.6 |
| G2 | 1.4 |
| G3 | 0.1 |
| G5 | 0.6 |
Example
Consider a DEG for 8 genes {G1 to G8}, where the functional pathway “PATH_A” contains 4 genes {G1, G2, G3, G4}. With the readings on the right, please calculate the enrichment score of PATH_A on this array.
| Gene | Log2FC |
|---|---|
| G1 | 2.3 |
| G2 | 0.7 |
| G3 | 2.6 |
| G4 | 1.4 |
| G5 | 1.2 |
| G6 | 0.1 |
| G7 | 0.3 |
| G8 | 0.6 |
Example
| Rank | Gene | Hit/Miss | Score |
|---|---|---|---|
| 1 | G3 | Hit | 2.6/7 |
| 2 | G1 | Hit | 2.3/7 |
| 3 | G4 | Hit | 1.4/7 |
| 4 | G5 | Miss | -1/4 |
| 5 | G2 | Hit | 0.7/7 |
| 6 | G8 | Miss | -1/4 |
| 7 | G7 | Miss | -1/4 |
| 8 | G6 | Miss | -1/4 |
- Score deducted if the gene is not in the pathway 1/(8-4)
- Normalized term (\(N_R\)) if the gene is in the pathway: 2.6+2.3+1.4+0.7=7
- ES
Example 2
Consider a DEG for 8 genes {G1 to G8}, where the functional pathway “PATH_B” contains 5 genes {G1, G2, G3, G4, G5}. With the readings on the right, please calculate the enrichment score of PATH_A on this array.
| Gene | Log2FC |
|---|---|
| G1 | -2.0 |
| G2 | -0.1 |
| G3 | -2.3 |
| G4 | -1.7 |
| G5 | -0.9 |
| G6 | 0.1 |
| G7 | 0.3 |
| G8 | 0.6 |
Example 2
| Rank | Gene | Hit/Miss | Score |
|---|---|---|---|
| 1 | G8 | Hit | -1/3 |
| 2 | G7 | Hit | -1/3 |
| 3 | G6 | Hit | -1/3 |
| 4 | G2 | Miss | 0.1/7 |
| 5 | G5 | Hit | 0.9/7 |
| 6 | G4 | Miss | 1.7/4 |
| 7 | G1 | Miss | 2/4 |
| 8 | G3 | Miss | 2.3/4 |
- Score deducted if the gene is not in the pathway: 1/(8-5)
- Normalized term (\(N_R\)) if the gene is in the pathway 2+0.1+2.3+1.7+0.9=7
- ES
Comparison of ORA and GSEA
ORA
- A hard threshold
- Provides a list of interested genes instead of DEG result
GSEA
- A soft threshold
- Needs a DEG result
- Computationally expensive
- Can determine the regulation direction
Different results in ORA and GSEA might implies subtle difference at the boundary of arbitrary cut-off (i.e. some genes with p-value like 0.051)
Biological Networks
Complexity of Interactions
In reality, proteins can interact together, and it’s more complex than what we’ve seen in KEGG.
Reasons
Proteins could interact with each other in other pathways (e.g., some interactions can be related to insulin resistance but not insulin secretion, so KEGG does not show it)
Scientific advancements and experiments:
- Gene co-expression
- Co-occurrence
Getting Networks
Getting Protein-Protein Interaction (PPI) Network
STRING: A database to document interactions between proteins
- Interactions:
- Physical and functional associations
- Sources:
- Computational prediction
- Knowledge transfer between organisms
- Databases
Definition of the Network
\[\begin{equation*} \begin{split} G&=(V,E) \\ V&=\text{All the nodes (Proteins)}\\ E&=\{(s,e)\} \quad \text{All the edges from s to e (interactions between two)} \end{split} \end{equation*}\]
Importance of a Node
Local Community of a Network
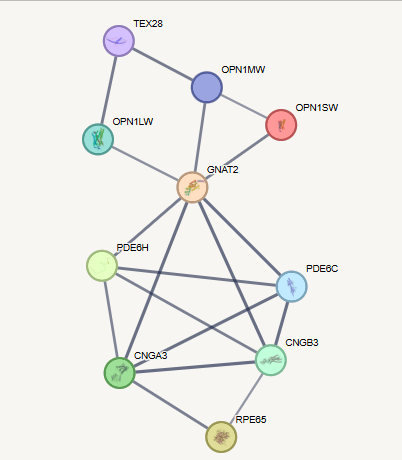
Clustering Coefficient
- How many friends of friends are also my friends (Triangles)
\[\begin{equation*} \begin{split} \text{Clustering coefficient(i)}&=\frac{L_i}{\frac{k_i(k_i-1)}{2}}\\ &=\frac{2L_i}{k_i(k_i-1)} \end{split} \end{equation*}\]
- \(L_i\): Number of triangles including node \(i\)
- \(k_i\): Number of nodes connected to node \(i\)
Clustering coefficient of:
- GNAT2: \(\frac{7*2}{7*6}=0.33\), TEX28: \(0\)
Why They Are Important
Degree
- Importance of a protein
- Identifying potential drug targets
Clustering Coefficient
- Identifies potential clusters (sub-functions)
- Identifying potential drug targets
Application of PPI
- Exploring disease mechanisms
- Drug discovery
Wrap Up
- Connecting sequencing results to more informative clinical information:
- Enrichment in ChIP-Seq and DEG
- PPI